| oligos_5-8nt_m1_shift9 (oligos_5-8nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_5-8nt_m1; m=0 (reference); ncol1=14; shift=9; ncol=23; ---------wwgACACGTGTcww
; Alignment reference
a 0 0 0 0 0 0 0 0 0 133 126 97 324 19 433 25 23 19 27 53 92 153 143
c 0 0 0 0 0 0 0 0 0 93 93 81 64 411 6 415 7 12 10 27 197 98 99
g 0 0 0 0 0 0 0 0 0 100 97 203 27 11 11 7 415 7 415 65 81 91 94
t 0 0 0 0 0 0 0 0 0 144 154 89 55 29 20 23 25 432 18 325 100 128 134
|
| ABF2_MA0941.1_JASPAR_shift8 (ABF2:MA0941.1:JASPAR) |
 |
0.956 |
0.765 |
4.952 |
0.966 |
0.986 |
5 |
3 |
57 |
1 |
1 |
13.400 |
1 |
; oligos_5-8nt_m1 versus ABF2_MA0941.1_JASPAR (ABF2:MA0941.1:JASPAR); m=1/119; ncol2=13; w=12; offset=-1; strand=D; shift=8; score= 13.4; --------cayrACACGTgkt--
; cor=0.956; Ncor=0.765; logoDP=4.952; NsEucl=0.966; NSW=0.986; rcor=5; rNcor=3; rlogoDP=57; rNsEucl=1; rNSW=1; rank_mean=13.400; match_rank=1
a 0 0 0 0 0 0 0 0 231 308 191 274 699 0 999 0 19 19 218 213 229 0 0
c 0 0 0 0 0 0 0 0 308 231 268 120 177 840 0 917 19 19 59 130 229 0 0
g 0 0 0 0 0 0 0 0 231 231 191 403 103 0 0 0 942 19 664 287 229 0 0
t 0 0 0 0 0 0 0 0 231 231 350 202 21 159 0 82 19 942 59 371 312 0 0
|
| BZR1_MA0550.1_JASPAR_rc_shift9 (BZR1:MA0550.1:JASPAR_rc) |
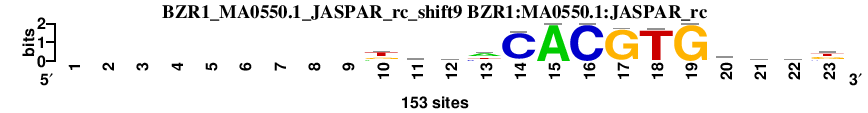 |
0.913 |
0.913 |
6.411 |
0.955 |
0.972 |
21 |
1 |
26 |
7 |
16 |
14.200 |
2 |
; oligos_5-8nt_m1 versus BZR1_MA0550.1_JASPAR_rc (BZR1:MA0550.1:JASPAR_rc); m=2/119; ncol2=14; w=14; offset=0; strand=R; shift=9; score= 14.2; ---------kkbhCACGTGkmkk
; cor=0.913; Ncor=0.913; logoDP=6.411; NsEucl=0.955; NSW=0.972; rcor=21; rNcor=1; rlogoDP=26; rNsEucl=7; rNSW=16; rank_mean=14.200; match_rank=2
a 0 0 0 0 0 0 0 0 0 33 28 16 72 0 153 0 0 4 0 12 51 24 0
c 0 0 0 0 0 0 0 0 0 0 22 43 39 142 0 153 0 0 0 26 52 27 30
g 0 0 0 0 0 0 0 0 0 39 61 53 0 5 0 0 147 2 153 66 20 47 60
t 0 0 0 0 0 0 0 0 0 81 42 41 42 6 0 0 6 147 0 49 30 55 63
|
| ATAREB1_M0256_1.02_CISBP_shift8 (ATAREB1:M0256_1.02:CISBP) |
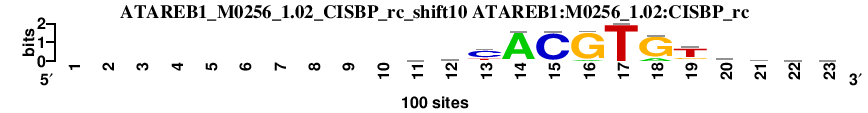 |
0.956 |
0.765 |
4.928 |
0.965 |
0.986 |
6 |
4 |
58 |
2 |
2 |
14.400 |
3 |
; oligos_5-8nt_m1 versus ATAREB1_M0256_1.02_CISBP (ATAREB1:M0256_1.02:CISBP); m=3/119; ncol2=13; w=12; offset=-1; strand=D; shift=8; score= 14.4; --------cayrACACGTgkt--
; cor=0.956; Ncor=0.765; logoDP=4.928; NsEucl=0.965; NSW=0.986; rcor=6; rNcor=4; rlogoDP=58; rNsEucl=2; rNSW=2; rank_mean=14.400; match_rank=3
a 0 0 0 0 0 0 0 0 23 31 19 27 70 0 100 0 2 2 22 21 23 0 0
c 0 0 0 0 0 0 0 0 31 23 27 12 18 84 0 92 2 2 6 13 23 0 0
g 0 0 0 0 0 0 0 0 23 23 19 41 10 0 0 0 94 2 66 29 23 0 0
t 0 0 0 0 0 0 0 0 23 23 35 20 2 16 0 8 2 94 6 37 31 0 0
|
| HY5_MA0551.1_JASPAR_shift8 (HY5:MA0551.1:JASPAR) |
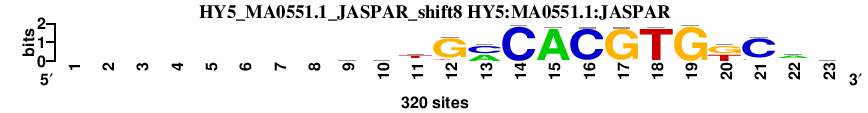 |
0.901 |
0.788 |
6.531 |
0.945 |
0.957 |
30 |
2 |
24 |
20 |
34 |
22.000 |
11 |
; oligos_5-8nt_m1 versus HY5_MA0551.1_JASPAR (HY5:MA0551.1:JASPAR); m=11/119; ncol2=16; w=14; offset=-1; strand=D; shift=8; score= 22; --------wwtGmCACGTGkCaw
; cor=0.901; Ncor=0.788; logoDP=6.531; NsEucl=0.945; NSW=0.957; rcor=30; rNcor=2; rlogoDP=24; rNsEucl=20; rNSW=34; rank_mean=22.000; match_rank=11
a 0 0 0 0 0 0 0 0 103 108 52 9 150 0 310 3 1 0 2 4 24 183 94
c 0 0 0 0 0 0 0 0 60 56 55 8 165 317 1 311 5 9 1 1 279 30 62
g 0 0 0 0 0 0 0 0 68 62 30 279 1 1 9 5 311 1 317 165 8 55 56
t 0 0 0 0 0 0 0 0 89 94 183 24 4 2 0 1 3 310 0 150 9 52 108
|




